Parallel in vivo analysis of large-effect autism genes implicates cortical neurogenesis and estrogen in risk and resilience (Willsey et al., Neuron, 2021)
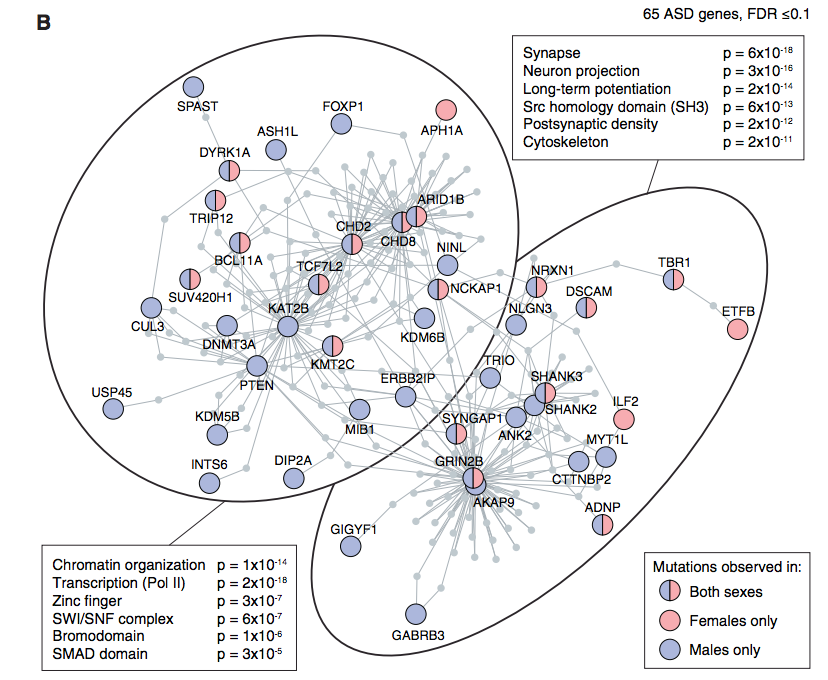
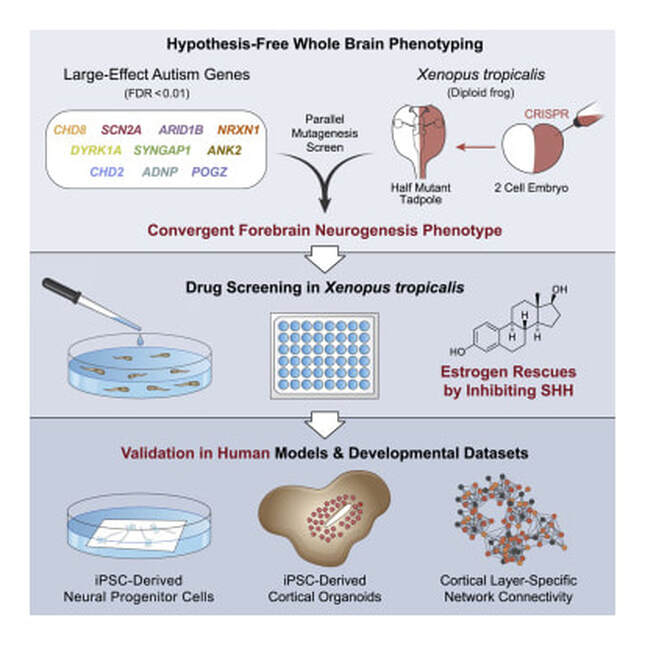
Gene Ontology analyses of autism spectrum disorders (ASD) risk genes have repeatedly highlighted synaptic function and transcriptional regulation as key points of convergence. However, these analyses rely on incomplete knowledge of gene function across brain development. Here we leverage Xenopus tropicalis to study in vivo ten genes with the strongest statistical evidence for association with ASD. All genes are expressed in developing telencephalon at time points mapping to human mid-prenatal development, and mutations lead to an increase in the ratio of neural progenitor cells to maturing neurons, supporting previous in silico systems biological findings implicating cortical neurons in ASD vulnerability, but expanding the range of convergent functions to include neurogenesis. Systematic chemical screening identifies that estrogen, via Sonic hedgehog signaling, rescues this convergent phenotype in Xenopus and human models of brain development, suggesting a resilience factor that may mitigate a range of ASD genetic risks.
Gene Ontology analyses of autism spectrum disorders (ASD) risk genes have repeatedly highlighted synaptic function and transcriptional regulation as key points of convergence. However, these analyses rely on incomplete knowledge of gene function across brain development. Here we leverage Xenopus tropicalis to study in vivo ten genes with the strongest statistical evidence for association with ASD. All genes are expressed in developing telencephalon at time points mapping to human mid-prenatal development, and mutations lead to an increase in the ratio of neural progenitor cells to maturing neurons, supporting previous in silico systems biological findings implicating cortical neurons in ASD vulnerability, but expanding the range of convergent functions to include neurogenesis. Systematic chemical screening identifies that estrogen, via Sonic hedgehog signaling, rescues this convergent phenotype in Xenopus and human models of brain development, suggesting a resilience factor that may mitigate a range of ASD genetic risks.